Gene expression profiling provides an affordable approach to examine differential gene expression analysis between groups of samples, such as various treatments, time-points, or disease versus control samples.

Gene Expression Profiling

Why analyse gene expression?

A sensitive indicator of changes in biological processes
Our ISO-accredited service includes all novel features: Unique Molecular Identifiers (UMIs), identification of antisense transcripts, and can handle a broad range of input RNA, starting from 10ng. All protein-coding (poly-A containing) transcripts are consistently and accurately represented.
For whole blood analysis, we offer globin reduction that removes the globin transcripts originating from erythrocytes, so you reduce the sequencing capacity that is required per sample with 30 to 40%. The removal of ribosomal RNA is not necessary, since these transcripts do not contain a poly-A tail. For custom transcript removal or targeted RNA seq-approaches, contact our scientific support team.
Data analysis
Our experienced bioinformaticians generate quality assured data-sets or deliver publication-ready results through our validated pipelines. For more demanding projects our experts work closely with you to provide a fully custom analysis.
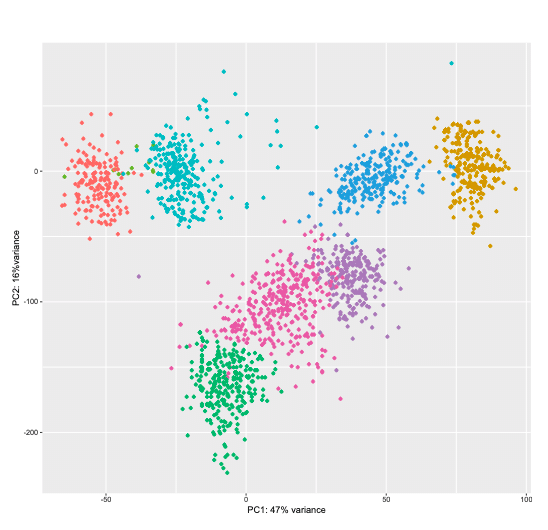
Principle component analysis, gene ontology enrichment and hierarchical clustering are examples of our extensive data analysis options for gene expression profiling.
Structural variant analysis
To determine structural variation such as isoforms, splice variants and gene-fusions we advise a sequencing depth of 50 – 100 million reads per sample.
We have summarized key information about our gene expression profiling service into a service specification sheet.
Let's get the conversation started for your next NGS project
Please either fill in this form or email us directly at info@genomescan.nl
and we will get in touch with you to discuss your requirements





