The EpiRestore™ service restores heavily degraded DNA so it can be analyzed using Illumina’s MethylationEPIC micro-array. Moreover, our modified hybridization protocol allows you to start with as little as 200 ng of DNA. You can harvest a wealth of data of previously unattainable patient cohorts.
EpiRestore™

Why use EpiRestore™?

DNA repair service
The advantages of EpiRestore™
- Applicable for sample cohorts with low DNA integrity, such as paraffin blocks or Guthry cards
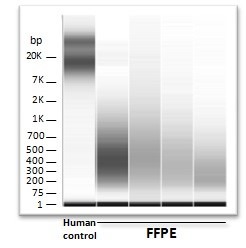
- Restores DNA that is heavily degraded down to average lengths of 300 basepairs
- Applicable for low amounts (200 ng) of genomic DNA
- Gives biologically meaningful data: the data-set highly resembles the sample as if it was fresh frozen and stored without fixative
- Excellent tool to screen large sample numbers due to cost-effective pricing and easy data-analysis.
Measuring epigenetic markers in paraffin embedded (FFPE) blocks is challenging. Fixation extensively damages and shreds DNA. The nucleotides of these DNA fragments also become altered by processes such as depurination and de-amination.
Our scientists have developed a proprietary DNA repair service that allows you to measure methylated and/or hydroxymethylated residues. This advanced method interrogates over 850,000 loci throughout the genome, selected by epigenetic experts. It delivers robust and highly reproducible results despite the very poor condition of the starting material.
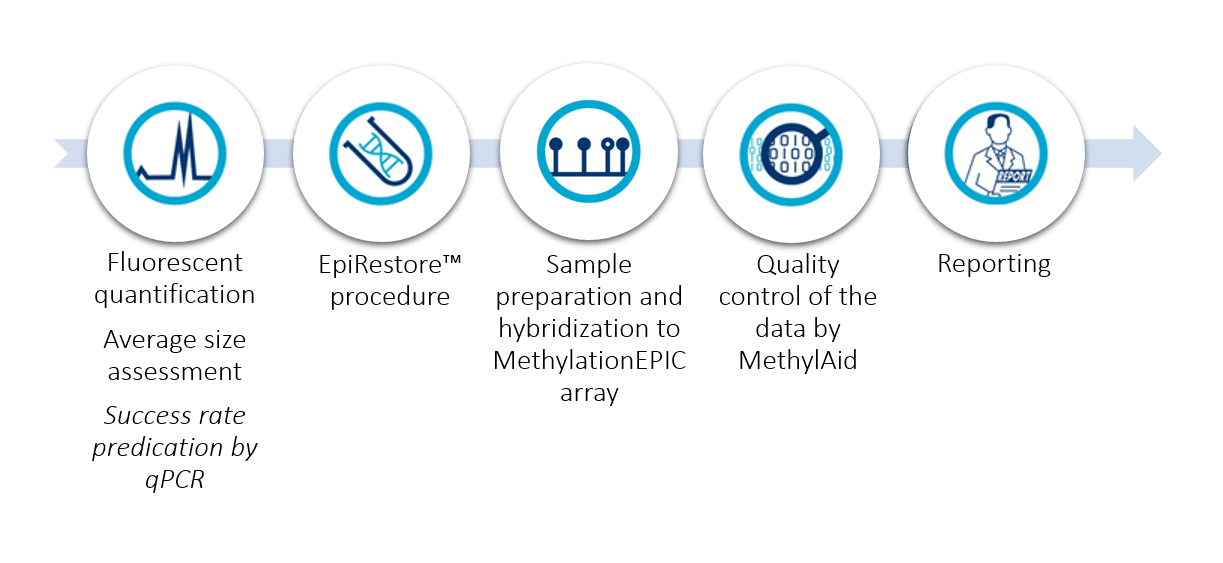
Service workflow
Quality checks
We perform quality checks throughout the whole procedure, to ensure that only good quality results will be reported. We deliver all micro-array quality specs already loaded into the intuitive open-source software package MethylAID (van Iterson, M. et al. Bioinformatics 2014). You can immediately assess the quality of the data by scrolling through the graphs. Each describe a quality aspect: sample-dependent as well as sample-independent controls such as call-rates, hybridization controls and bisulfite conversion controls.
Guarantee for highly degraded FFPE samples
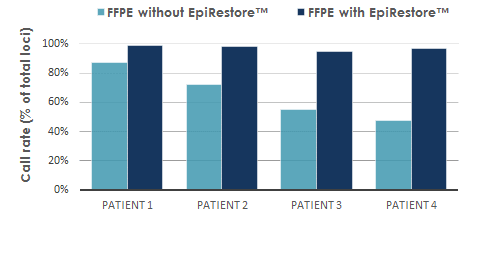
Our R&D scientists have processed many FFPE cohorts and have shown that we can extract biological meaningful data with great statistical certainty, even from highly degraded cohorts.
You retrieve the methylation status of 765,000 or more loci, from CpG sites selected by Epigenetics experts: a pan-enhancer and coding region view of the methylome, ready for use in epigenome-wide association studies on various human tissues.

The MethylationEPIC BeadChip contains >90% of the original 450K BeadChip content, allowing you to add FFPE sample data to previously analyzed cohorts.
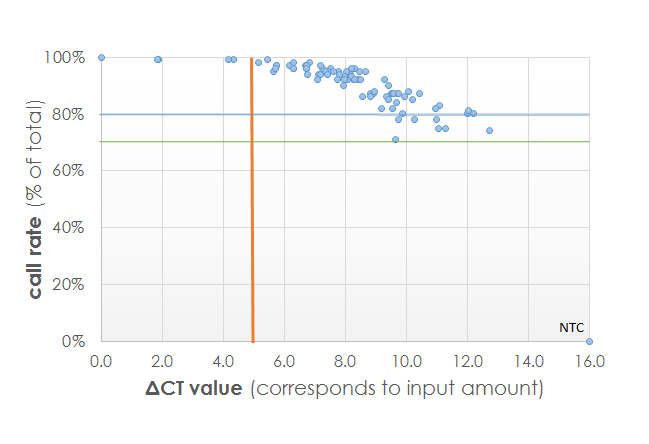
Service validation data
EpiRestore™ guarantee
For freshly isolated DNA, we routinely detect call rates of above 99%. We can guarantee to detect 95% of the loci with a p-value of p≤0.01). With our fine-tuned protocols we can start this procedure with 100 ng input or even less, in contrast to the 250 ng input recommended by Illumina. Contact technical support to inquire.
The results obtained by EpiRestore™ are depending on the quality of the DNA. We guarantee that at least 90% of the CpGs are detected at p<0.01 for a minimum of 80% of the samples. Thus, at least ~765,000 CpG loci that show differential methylation patterns in e.g. health and disease, are called with a high level of statistical certainty. This guarantee is valid for input amounts of at least 200 ng, enabling you to select only the regions of interest from the paraffin blocks.
| Application | Restoration
required |
Percentage of detected CpGs guaranteed | Min. no. of detected CpGs
(p ≤ 0.01) |
For number of samples |
| Methylation | No | 95 % | 807,500 | 100 % |
| FFPE Methylation | Yes | 90 % | 765,000 | 80 % |
| Hydroxymethylation | No | 95 % | 807,500 | 100 % |
| FFPE Hydroxymethylation | Yes | 90 % | 765,000 | 80 % |
Our scientists can give these guarantees because we have challenged our partners to send in the most degraded samples available. The most interesting cohort was especially selected for this purpose by a highly-regarded team of the Karolinska Institute. Even with fragment lengths of less than 300 bp (instead of the average DNA fragment size of 20,000 bp), the EpiRestore™ procedure delivered biologically relevant data, thereby fulfilling our GenomeScan guarantee.
Finding biomarkers with consortium partners
Biomarkers are characteristics by which a disease can be identified or monitored. They reflect past environmental exposures, predict disease onset or course, or determine a patient’s response to therapy. GenomeScan partners now with many (academic) institutes to find diagnostic and predictive biomarkers for cancer and other diseases, based on epigenetic markers on the DNA. We participate in multiple consortia. Visit our research collaborations page to learn about how to successfully apply for funding together with us.
Let's get the conversation started for your next NGS project
Please either fill in this form of email us directly at info@genomescan.nl
and we will get in touch with you to discuss your requirements