Getting the best transcriptomics results out of your FFPE cohort requires expertise. GenomeScan offers you a comprehensive, robust, and reproducible solution for FFPE RNA analysis.

FFPE RNAseq

Is FFPE RNA sequencing challenging?

How to ensure FFPE RNAseq reliability
Extracting gene-expression profiles from Biobanks
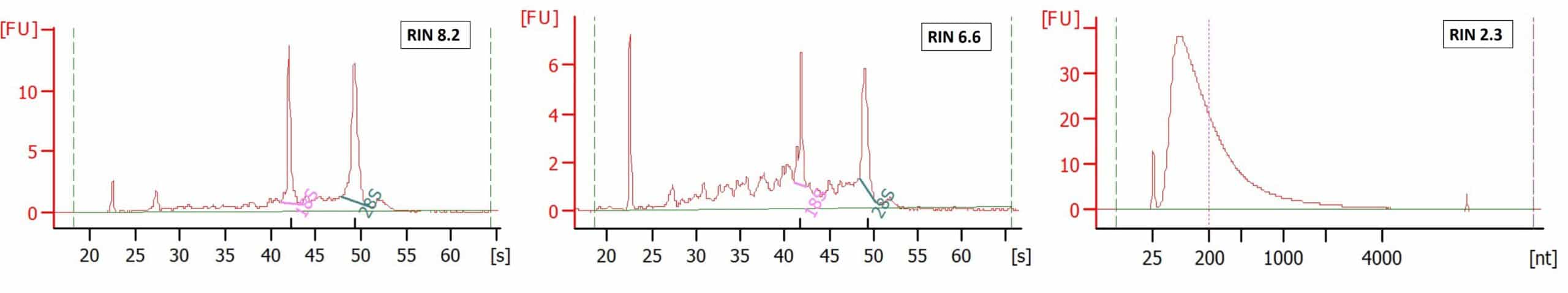
Biobanks harbor a wealth of gene-expression data of patients with a certain disease. The effects of treatment are documented and can be followed over time on molecular level. These biobanks generally consist of Formalin-Fixed Paraffin-Embedded (FFPE) tissue. High degree of RNA degradation of such FFPE cohorts is often hampering analyses by, for instance, leading to exclusion of key samples and/or essential time points. Factors that affect RNA quality are e.g. age of the blocks, treatment of the material before fixation, storage conditions. Also, formalin fixation damages RNA, causing extensive shredding and modification of nucleotides.
GenomeScan’s solution for FFPE RNA analysis
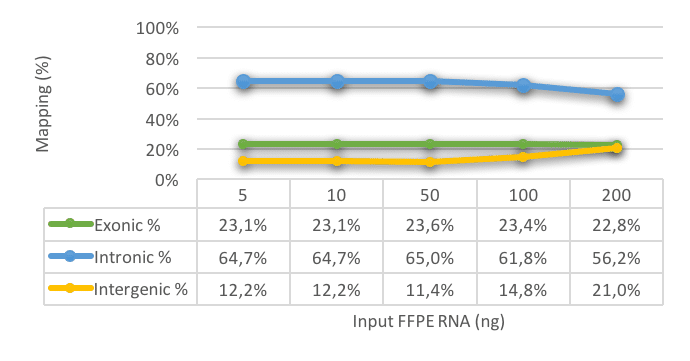
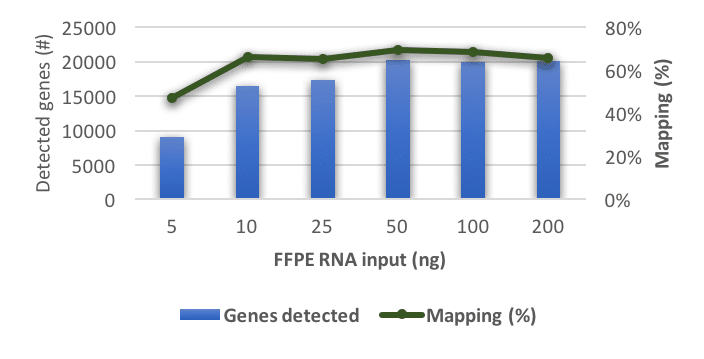
Our scientists have thoroughly studied the effects of RNA degradation and RNA yield on the accuracy and reliability of the transcriptomics data.
In validation studies performed with the library preparation chemistry of New England Biolabs (NEB) combined with Illumina sequencing, we have determined the parameters that are best suitable for monitoring the validity of the data. The figure, below on this page, shows that the mapping of FFPE material remains consistent, even with less input. Thus, gene-expression analysis of FFPE RNA is robust and reproducible in the range of 5 to 200ng, for this cohort.
Frequently asked questions
How does FFPE compare to Fresh Frozen material?
Studies generally include a comparison between Fresh Frozen (FF) material and FFPE. The goal is to show that data derived from various sources, will generate similar results.
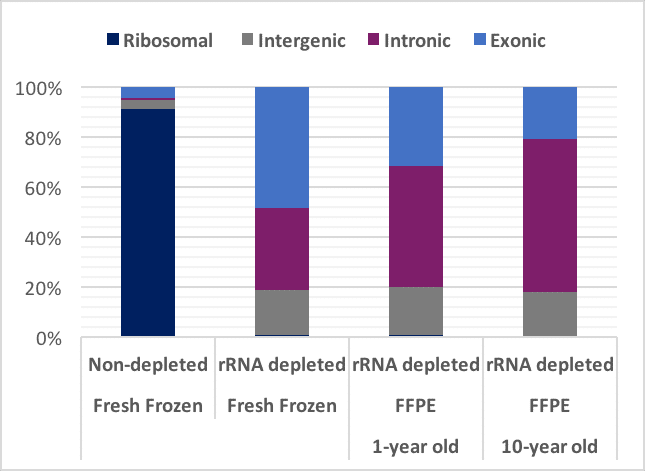
As elegantly shown by our partners of New England Biolabs (NEB), fixation will always lead to a certain change of the transcripts that you retrieve (see figure below). Factors that affect RNA quality are e.g. age of the blocks, treatment of the material before fixation, storage conditions. The GenomeScan entry QC will measure the extent of fragmentation of the RNA. The effect of other factors such as depurination will be measured in the small pilot study in which we perform the whole workflow. This will provide you direct and sample-specific information on the quality of your samples.
Figure: Effect of sample fixation on the read distribution. Figure adapted from the NEB rRNA depletion kit. Data generated using the Universal Human RNA Reference (UHR).
Service validation data
We have summarized key information about our FFPE RNAseq service into a service specification sheet.
Let's get the conversation started for your next NGS project
Please either fill in this form or email us directly at info@genomescan.nl
and we will get in touch with you to discuss your requirements.