Illumina’s Infinium MethylationEPIC v2.0 BeadChip allows you to interrogate over 935,000 methylation sites, selected by experts in the field. The results give an overview of the total methylome, which can be used for Epigenome-Wide Association Studies (EWAS) on various human tissues.
MethylationEPIC BeadChip
Why use MethylationEPIC BeadChip?

Our human methylation profiling service
The Infinium MethylationEPIC v2.0 BeadChip builds upon the genome-wide backbone of the Infinium MethylationEPIC v1.0 BeadChip, maintaining high backwards compatibility while adding new content informed by expert feedback and epigenetic evaluations of human cancers and cellular samples. Nonfunctional probes routinely filtered out in DNA methylation studies due to underlying single nucleotide polymorphisms (SNPs), cross-hybridization, and multimapping behavior were removed in the new version of the BeadChip, leaving space for more functional content identified by the epigenetics community. This array combines a unique wealth of data with low costs per sample and easy interpretation. Illumina’s MethylationEPIC BeadChip is an excellent tool for quantitative methylation measurement at single CpG level.
Content of the MethylationEPIC BeadChip
- Combination of Coding and Enhancer regions
- CpG sites outside of CpG islands
- Differentially methylated sites identified in tumor vs. normal
- Non-CpG methylated sites identified in human stem cells (CHH sites)
- miRNA promoter regions
Service workflow
Quality inspection
Our automated and robust methylation workflow results in an unprecedented high concordance between arrays. The correlation between technical replicates is generally above 0.99. That is why we guarantee to report at least 95% of all loci present on the MethylationEPIC BeadArray.
Highly reproducible data between both BeadChips and experiments is especially important when identifying small epigenetic changes or when multiple experiments must be combined, such as in cohort studies.
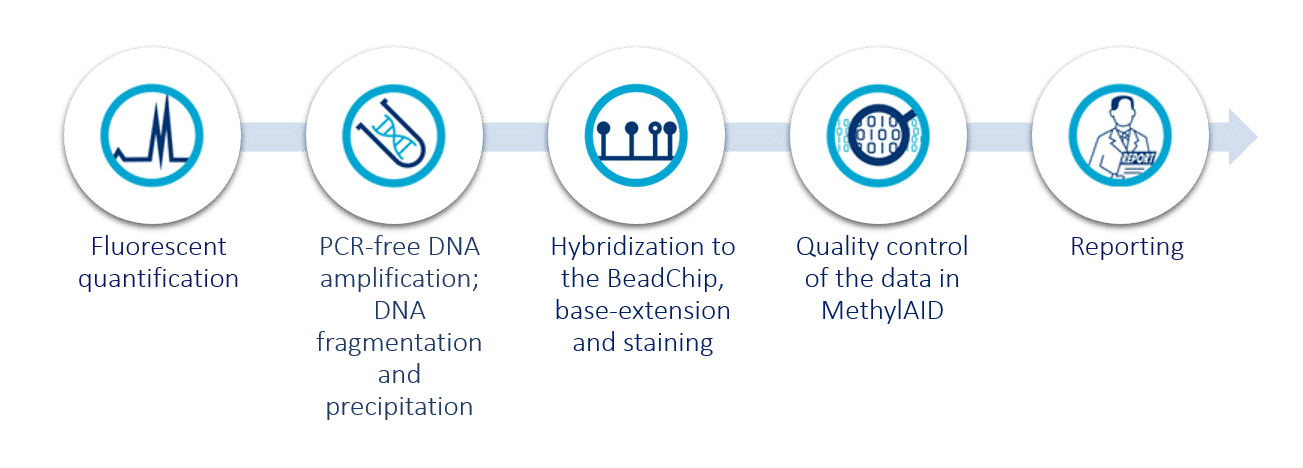
How it works
Infinium Methylation Assay Overview
The Infinium Methylation Assay provides quantitative array-based methylation measurement at the single-CpG-site level, offering high resolution for understanding epigenetic changes. Genome-wide methylation analysis capabilities make this assay highly suitable for studying the biological role of DNA methylation in normal and diseased cells.
Powerful Infinium Chemistry
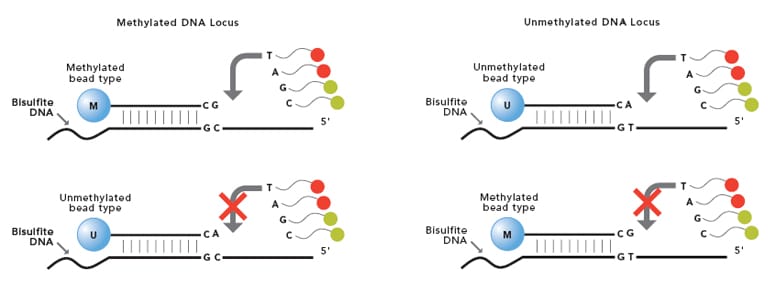
The Infinium Methylation Assay detects cytosine methylation at CpG islands based on highly multiplexed genotyping of bisulfite-converted genomic DNA. Upon treatment with bisulfite, unmethylated cytosine bases are converted to uracil, while methylated cytosine bases remain unchanged. The assay interrogates these chemically differentiated loci using two site-specific probes, one designed for the methylated locus (M bead type), and another for the unmethylated locus (U bead type). Single-base extension of the probes incorporates a labeled ddNTP, which is subsequently stained with a fluorescent reagent. The level of methylation for the interrogated locus can be determined by calculating the ratio of the fluorescent signals from the methylated vs. unmethylated sites.
We have summarized key information about our genome wide methylation analysis service into a service specification sheet.
Benefits of choosing GenomeScan
- Full service, including automated DNA isolation for FFPE
- Visualization of all quality metrics in MethylAID for direct assessment of samples and experiment performance
- QC measurements at multiple stages to tightly monitor sample quality
- Highly specialized researchers perform the experiments leading to unprecedented reproducibility
- High throughput and automation leads to cohort-friendly pricing
- Excellent scientific advice in experimental design
- One project manager that keeps you informed of the experiments at every stage of the project
Let's get the conversation started for your next NGS project
Please either fill in this form or email us directly at info@genomescan.nl
and we will get in touch with you to discuss your requirements.

