Transcription profiles give accurate images of the biological processes that influence cell fates. However, library construction can introduce bias at multiple steps. For example, PCR amplification increases the number of cDNA molecules to an amount sufficient for sequencing, but also stochastically introduces errors and amplifies different molecules with unequal probabilities.
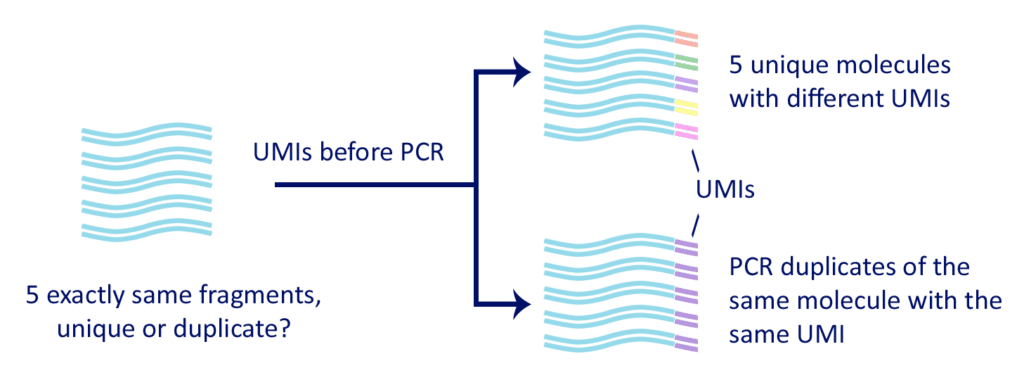
The sequencing reads that have been created from the same cDNA molecule by PCR are considered PCR duplicates. So, as a common practice at the data analysis stage, all but one identical reads are removed. This assumption may be flawed, especially when generating more data per sample, which increases the chance of eliminating identical reads that in fact originate from different cDNA molecules. Fu and colleagues showed that computational removal of PCR duplicates based only on their mapping coordinates introduces a substantial bias.
Four major reasons why UMIs improve transcriptomics data
The accuracy of gene-expression profiling experiments can increase drastically with the inclusion of unique molecular identifiers or UMIs.
- Especially in low frequency transcripts, the removal of duplicates by UMIs lead to more statistically significant findings.
- UMIs remove bias caused by variation in transcript length. A shorter gene is more likely to give rise to identical RNA-seq reads than a longer gene with the same transcript level, simply because of its size.
- UMIs improve datasets when duplicate removal is not an option. Especially for small RNA-seq reads such as microRNAs (miRNAs) or PIWI-interacting RNAs (piRNAs) standard duplicate removal is not an option, since many transcripts are identical.
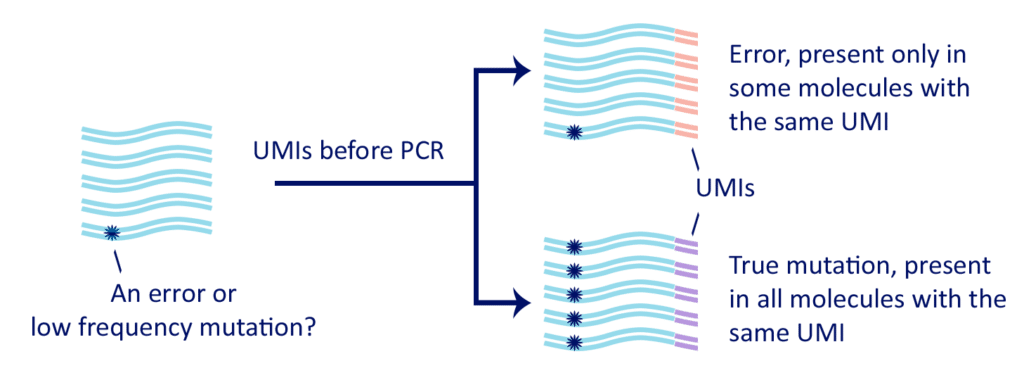
- Correction of PCR and sequencing errors. Duplicate reads are very useful: reads that are true duplicates can be used for data-correction. Especially when RNA-seq is used for qualitative analysis, such as SNP detection, inclusion of UMIs lead to a substantial improvement of the accuracy.
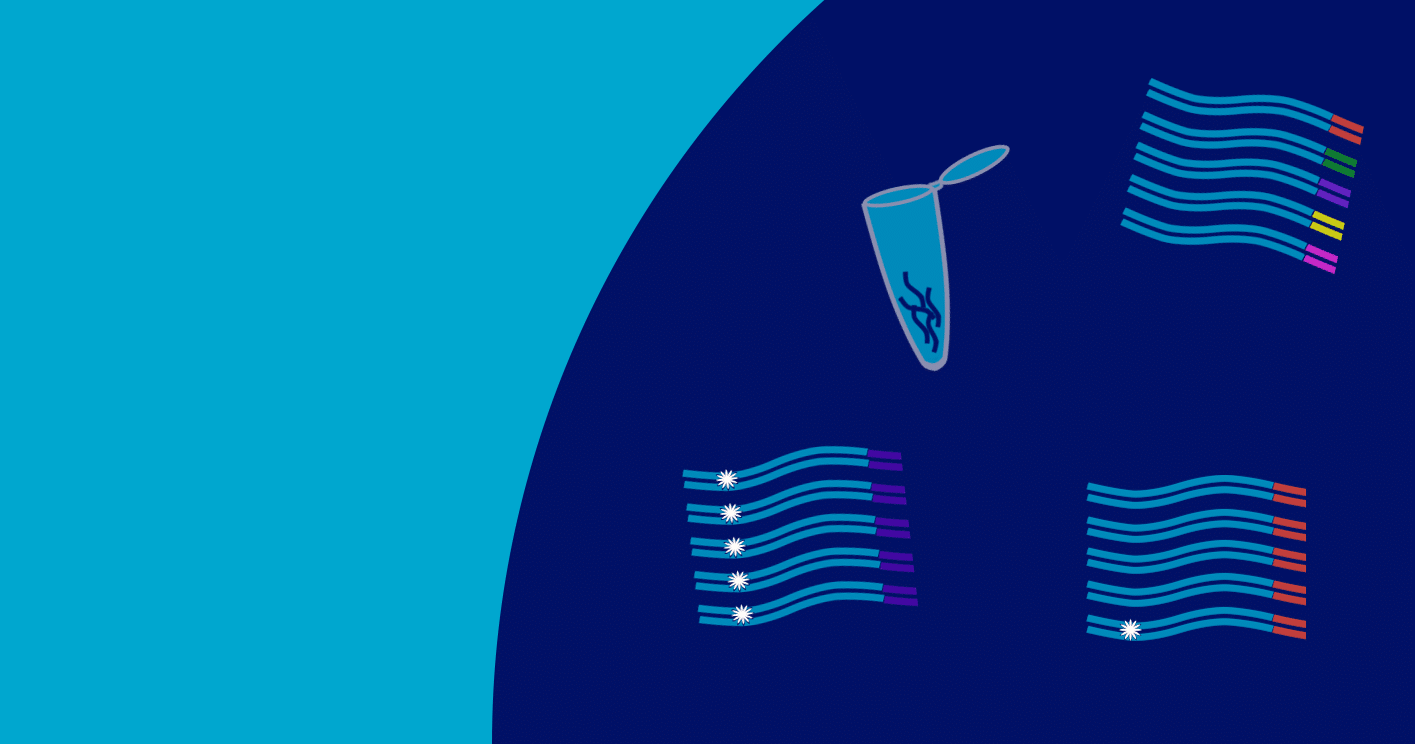
A less frequent but still important phenomenon is that identical small RNAs can be produced from multiple genomic loci. Since they map to the exact same genomic location, they are considered to be duplicates when in fact they arise from distinct sites in the genome.
Overall, UMIs contribute to a higher data-accuracy of your gene-expression profiling experiment. These technical innovations ensure a better representation of the transcripts in your samples. Inclusion of the appropriate bioinformatic handling of UMIs will simplify downstream analysis and interpretation of the data.
UMIs are part of the GenomeScan RNAseq portfolio
- Gene-expression profiling
- Total transcriptome
- Small RNAseq*
- Ultra-low input transcriptomics
(*UMIs implemented in 2020)
Do you work with any of the above methods? Tell us about your project and let’s see how your experiment can benefit from the use of UMIs.





Totally agree with you Zoraide, the appreciation of UMI’s is still not fully recognized. Thanks for sharing,
Kind regards,
Leendert Vermeulen
Roche diagnostics